Research in my lab is highly inter-disciplinary and spans across the fields of physiology, biochemistry, environmental biology, aquatic toxicology, and analytical chemistry. Specifically, I employ omics methods to understand the toxicological actions and broader ecological impacts of aquatic pollution and environmental stress.
Protein and metabolite profiling involves the large-scale identification of the proteome and metabolome for the purpose of differential comparison using full-scan mass spectrometry. Due to the randomized nature of this approach, protein and metabolite profiling can enable simultaneous understanding of an organism’s health, discovery of biomarkers specific to disease or environmental exposure, characterization of disease or toxicological mechanisms, and quantitative measurement of specific biomarkers within a single investigation.
 Environmental ‘omics is a growing field of interest, and protein and metabolite profiling can be integrated with gene transcription and chemical contaminant analyses so that an entire picture of an organism’s health can be understood by comparing an exposed or stressed organism to a healthy control organism. We have successfully applied this technique to studies involving the health of wild fish in the Athabasca Oil Sands, the Great Lakes Areas of Concern, and the effects of specific contaminants to model organisms (goldfish and rainbow trout) through laboratory exposures.
Environmental ‘omics is a growing field of interest, and protein and metabolite profiling can be integrated with gene transcription and chemical contaminant analyses so that an entire picture of an organism’s health can be understood by comparing an exposed or stressed organism to a healthy control organism. We have successfully applied this technique to studies involving the health of wild fish in the Athabasca Oil Sands, the Great Lakes Areas of Concern, and the effects of specific contaminants to model organisms (goldfish and rainbow trout) through laboratory exposures.
Specifically, I use liquid-chromatography tandem mass spectrometry to characterize and identify thousands of peptides and metabolites present in complex biological samples (biofluids and tissues) which I obtain from different tissues of teleost fish and other aquatic organisms. Much of my research focuses upon the meta-analysis of large datasets and the functional annotations of proteins using bioinformatics software and multivariate statistical analyses. I am always on the look-out for innovative analytical methods which can enhance the acquisition or interpretation of environmental ‘omics datasets.
The primary goal of my research is to create a database or library of plasma protein and metabolite expression signatures (PMES) that are specific to different classes of biologically active chemicals (for example, endocrine disruptors like ethinyl estradiol and bisphenol A, or heavy metals like mercury and cadmium, which can then be used as a diagnostic tool to identify when an organism has been exposed to a particular contaminant.
Blood Plasma for Monitoring Wildlife
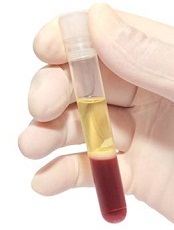 Blood plasma is the pale yellow liquid component of blood that holds the blood cells in suspension. Plasma has long been used in medicine for diagnosis and treatment of disease. It is mostly water (up to 95%) and contains dissolved proteins, metabolites, electrolytes, clotting factors, hormones, dissolved gases, and glucose. Plasma is involved in signaling, transport, protein storage, and waste clearance.
Blood plasma is the pale yellow liquid component of blood that holds the blood cells in suspension. Plasma has long been used in medicine for diagnosis and treatment of disease. It is mostly water (up to 95%) and contains dissolved proteins, metabolites, electrolytes, clotting factors, hormones, dissolved gases, and glucose. Plasma is involved in signaling, transport, protein storage, and waste clearance.
Plasma can be sampled non-lethally from wildlife with minimal impact to the individual, and thus could be the ideal biofluid for wildlife monitoring of sensitive populations. Plasma also contains both metabolites and proteins, is unencumbered by connective tissues which simplifies sample preparation, and as a circulating body-fluid, the molecules present in plasma represent most organs and systems. Plasma could provide information to construct complete mechanistic pathways, and thus it can indicate whole organism health.
However, plasma is challenging because of the variable range of responses between individuals and it’s function is largely unexplored in wildlife and model organisms used in ecotoxicology. Thus, as a secondary and more long-term goal, I would like to better understand the function of plasma in vertebrate organisms so that it could be used for non-lethal health monitoring of wildlife populations. Further to that, I would like to determine what classes of protein and metabolite molecules are expressed in blood plasma when specific organ systems or signaling pathways become challenged by their environment, and how long it would take for plasma to present a PMES response after the challenge has occurred.
